
IndraLab
INDRA Database

The INDRA (Integrated Network and Dynamical Reasoning Assembler) Database is a framework for creating, maintaining, and accessing a database of content, readings, and statements. This implementation is currently designed to work primarily with Amazon Web Services RDS running Postrgres 14+. Used as a backend to INDRA, the INDRA Database provides a systematic way of scaling the knowledge acquired from other databases, reading, and manual input, and puts that knowledge at your fingertips through a direct Python client and a REST api.
Database Statistics (As of Jan 2026)
Statements by Entity Type
This diagram shows how often different types of entities appear together in INDRA statements,
providing a high-level view of the interaction landscape in the database.
“Other” includes organisms, anatomical regions, and cellular locations.
Top interactions include:
| Interaction | Count |
|---|---|
| {{ labelMap[p.source]?.join(' ') || p.source }} → {{ labelMap[p.target]?.join(' ') || p.target }} | {{ formatNumber(p.value) }} |
Sources
The INDRA Database currently integrates and distills knowledge from several different sources, both biology-focused natural language processing systems and other pre-existing databases
Source Details
| Source Name | Type | Evidence Count |
|---|---|---|
| {{ source.name }} {{ source.name }} | {{ source.type }} | {{ source.count.toLocaleString() }} |
| Loading statistics... | ||
Content Insights
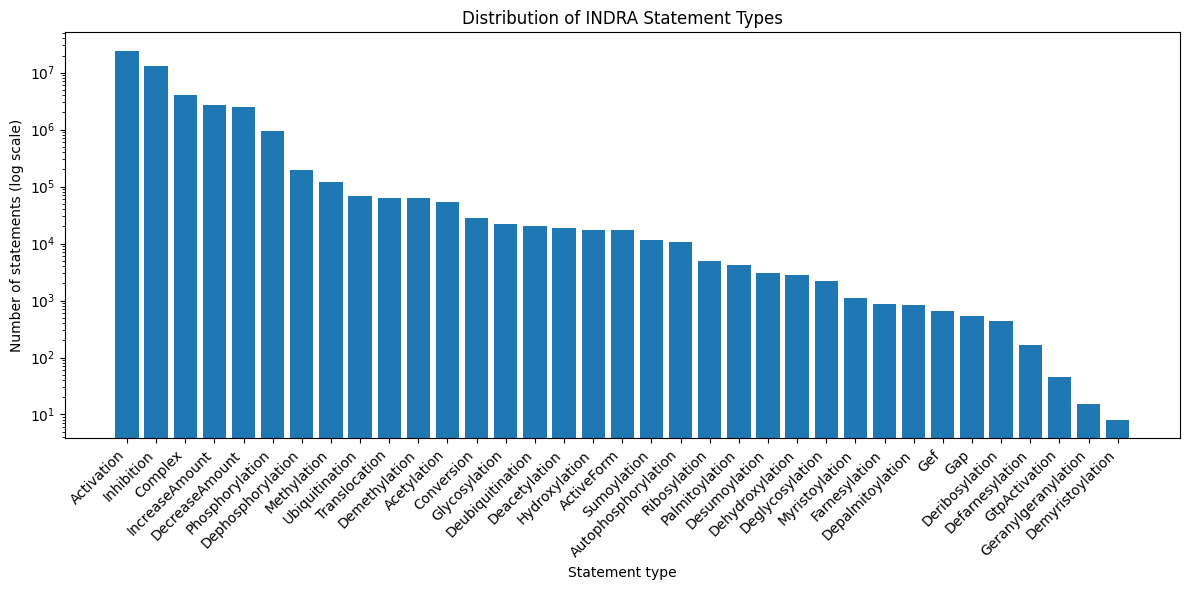
Statement Type Distribution
The statement type distribution shows a diversity of statement relations across 45 distinct types (e.g. activation, phosphorylation).
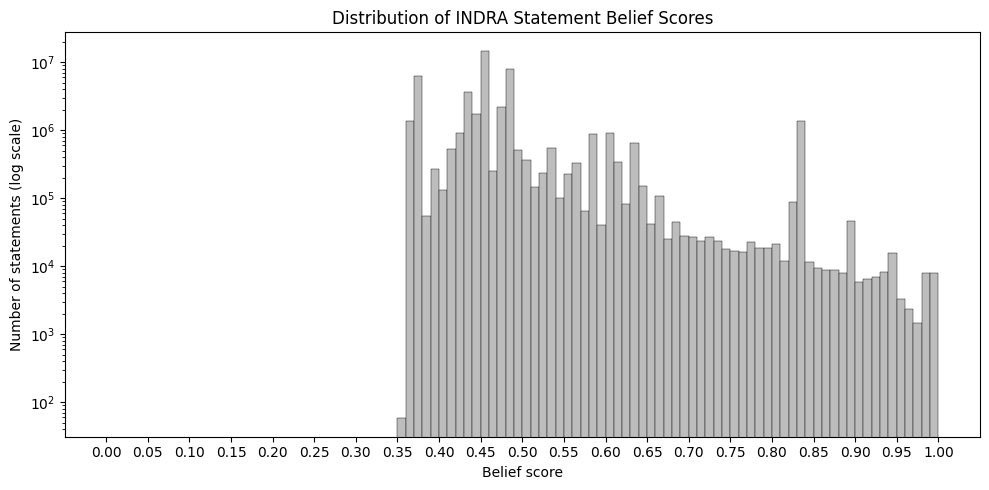
Belief Score Distribution
Belief scores are computed using INDRA’s HybridScorer, which combines a data-driven CountsScorer with a prior-based SimpleScorer. For statements whose evidence sources were observed during training, the CountsScorer estimates reliability. Support for more specific statements is propagated to their corresponding generalized statements, increasing the belief scores of the latter.
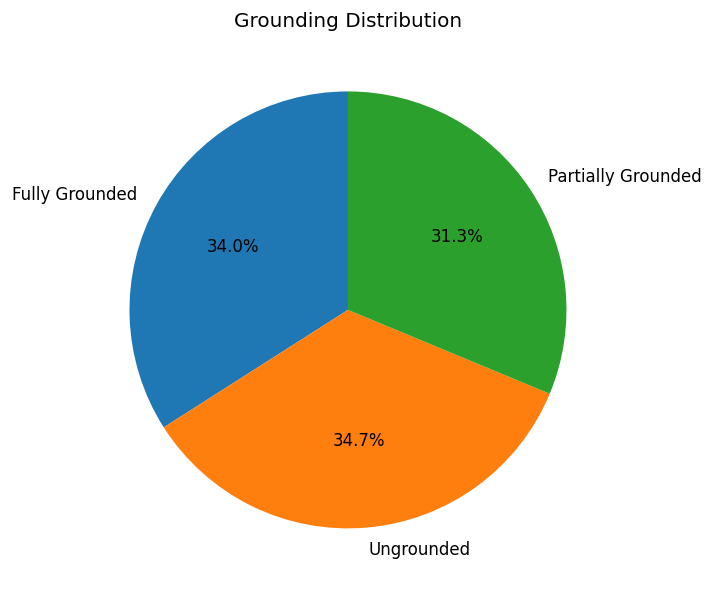
Grounding Distribution
After collecting and assembling all sources, the resulting corpus indexed by the database contains a total of 47,956,726 unique INDRA Statements. Of these, nearly 65.2% referred to at least one ungrounded entity. While statements about protein complexes can involve more than two entities, the vast majority 98.3% involved exactly two entities.
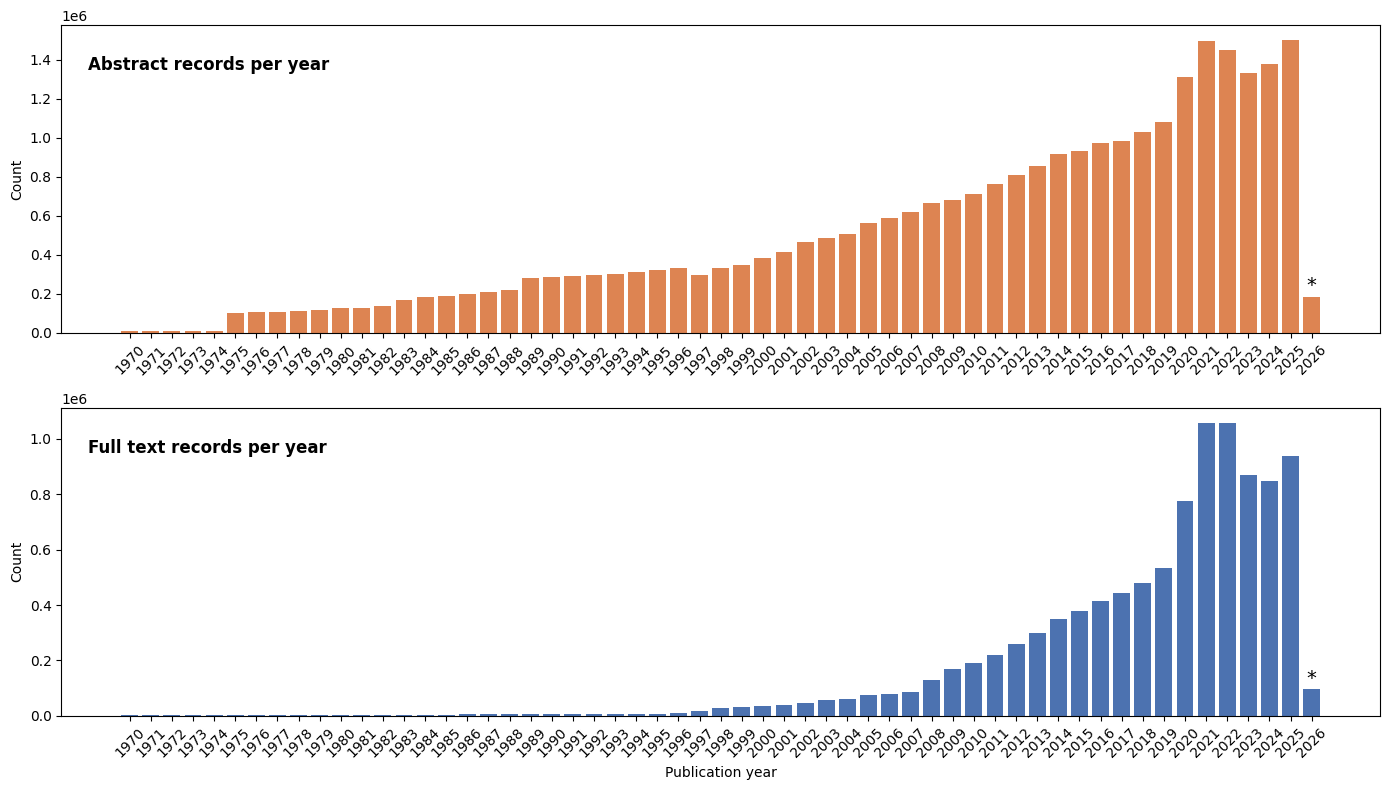
Paper Trends (Abstracts vs Fulltext)
A steady increase from year 1970 to 2024 exhibits a growing trend in the number of papers over time. * Data for 2026 reflect content by Jan 2026 and do not represent a complete calendar year. Before 1970, starting 1780, there are 52,779 abstract and 226,925 full text records in the database.
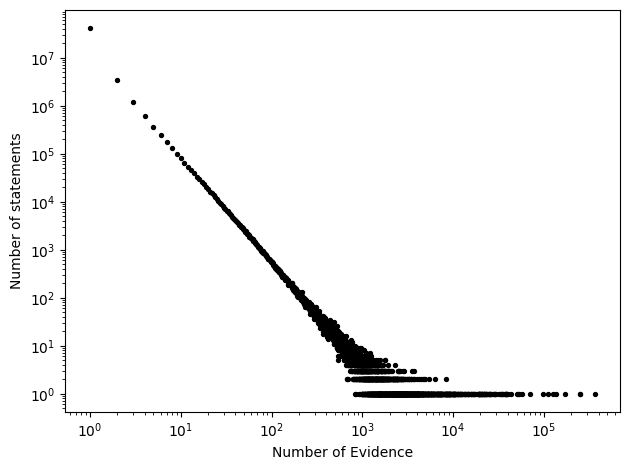
Evidence vs Statement Count
The scatter plot shows a strong inverse relationship between the number of evidence and the number of statements on a log–log scale. Most statements are supported by only a small number of evidence, while a very small number of statements aggregate a disproportionately large amount of evidence, forming a clear long-tailed distribution.
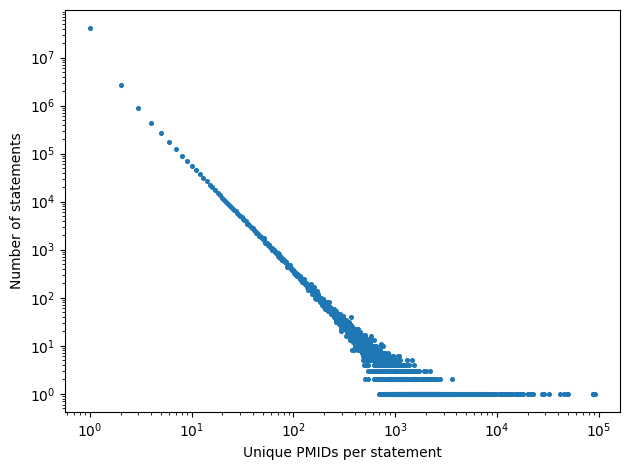
PMID vs Statement Count
The plot shows a strong inverse relationship between the number of unique PMIDs per statement and the number of statements on a log–log scale. The distribution is highly skewed, with most statements supported by only a few PMIDs, while a small fraction of statements accumulate evidence from hundreds to tens of thousands of publications.
Gene-Disease MeSH Distributions
These plots show what Medical Subject Headings disease terms are associated with publications from which statements about a given gene are described. For instance, INDRA Statements about EGFR are most commonly extracted from lung cancer and breast cancer publications. In contrast, Statements on SNCA are primarily from Parkinson's disease literature.
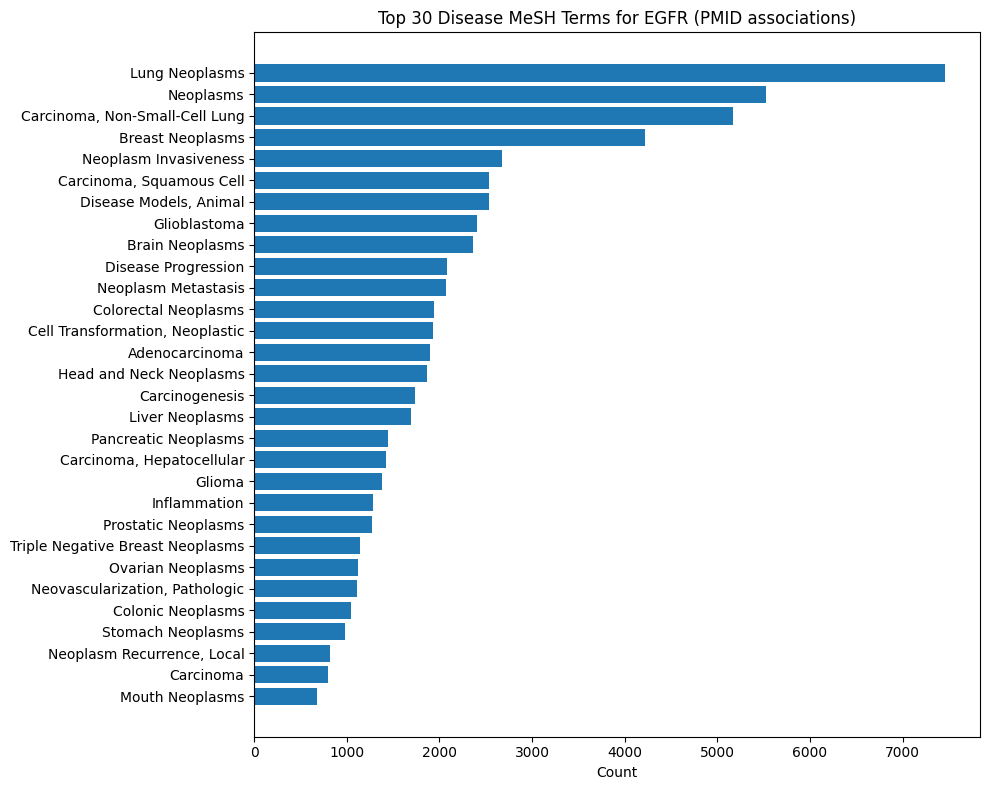
EGFR
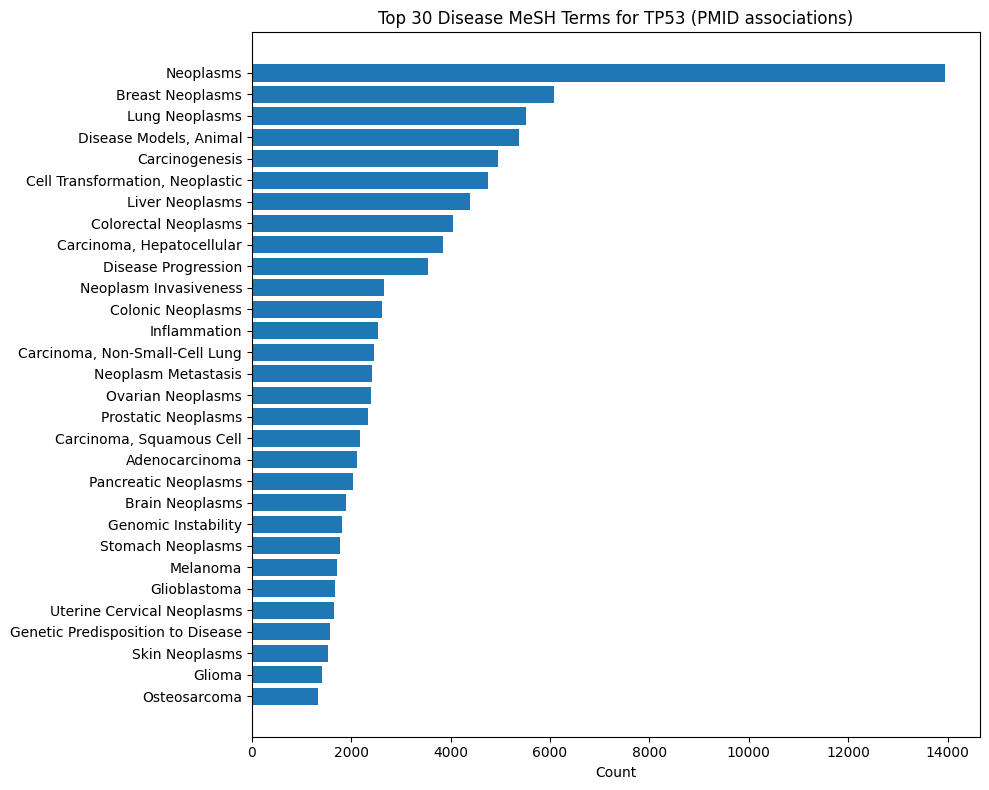
TP53
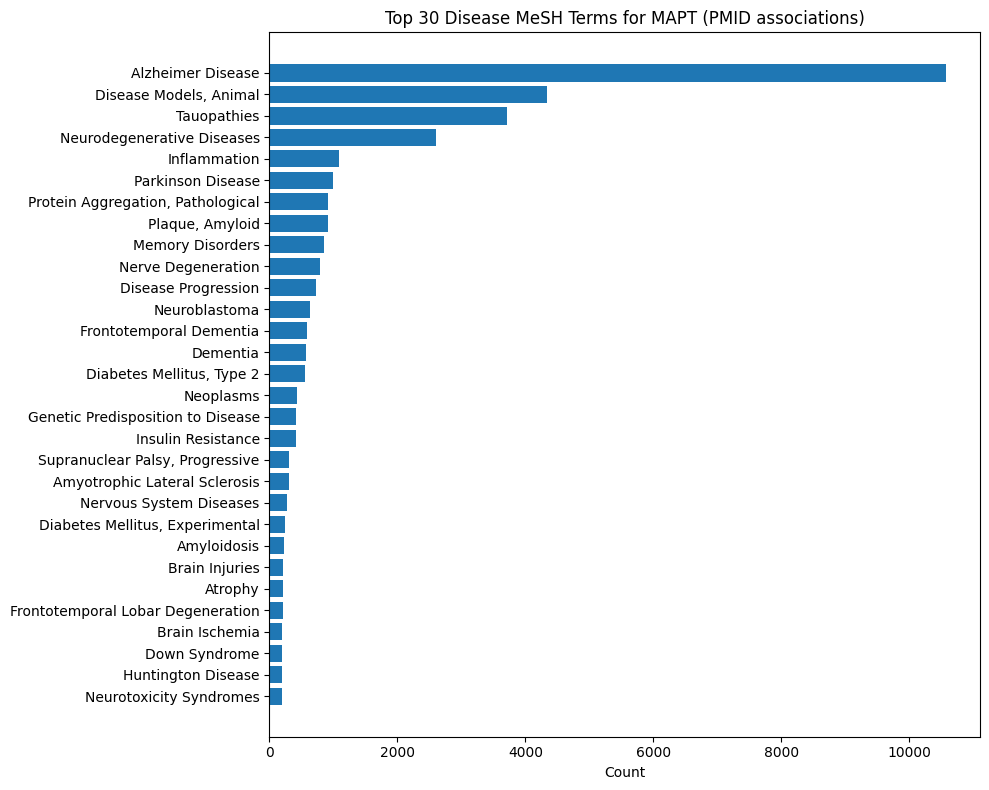
MAPT
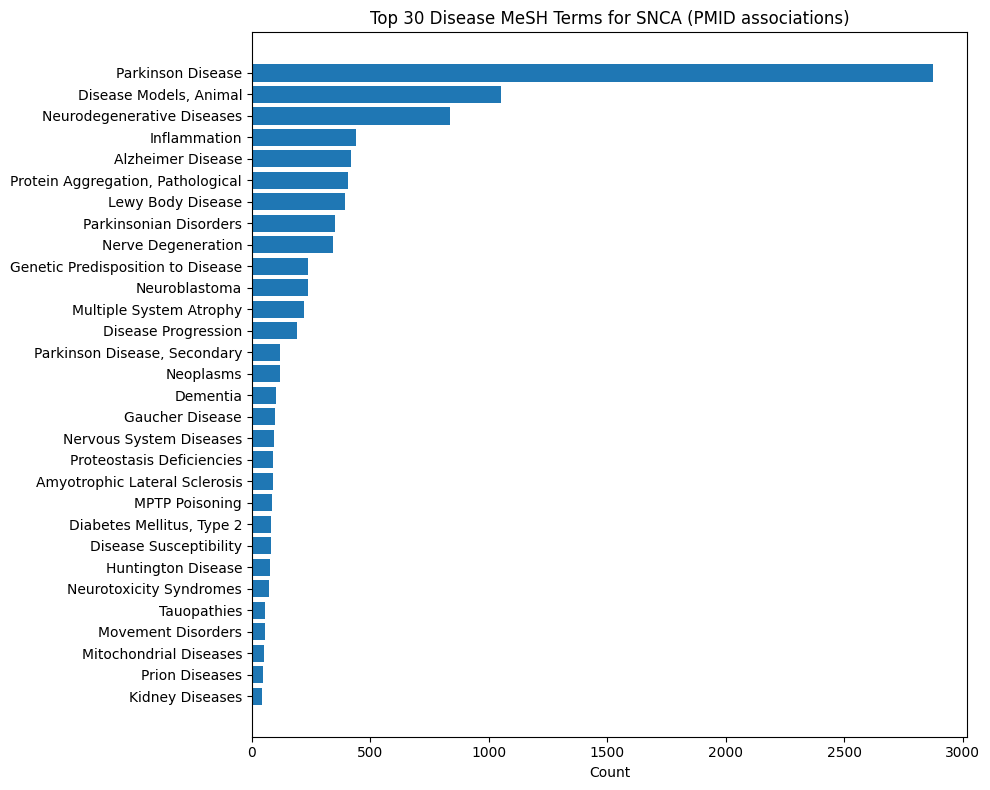
SNCA
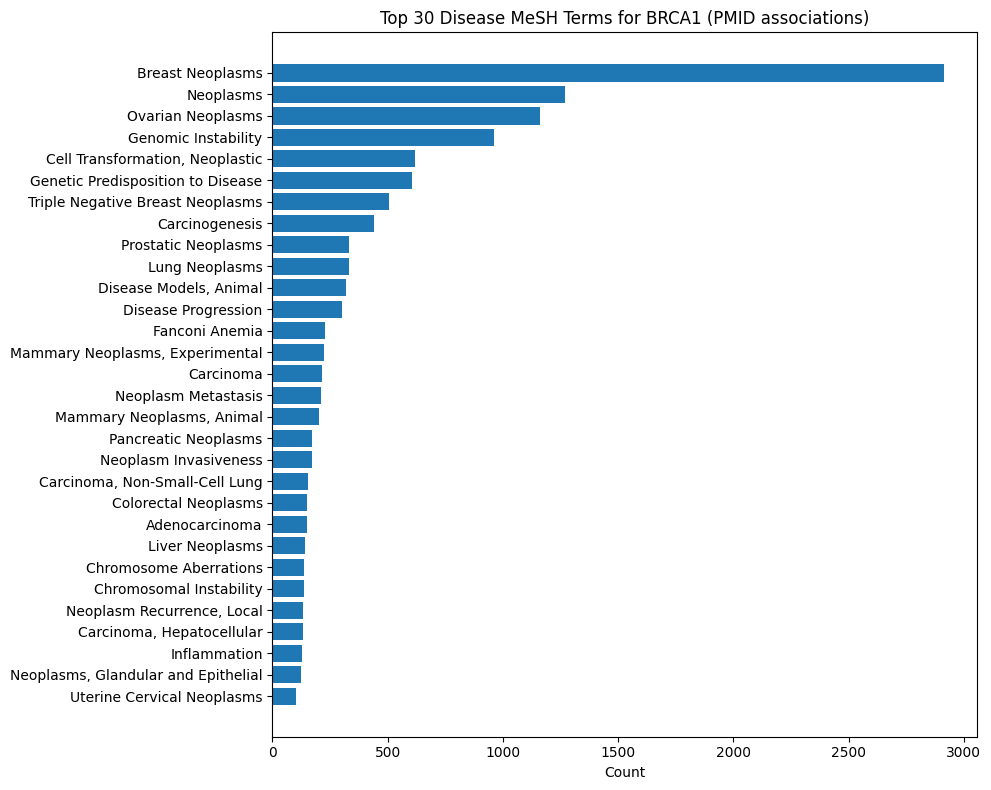
BRCA1
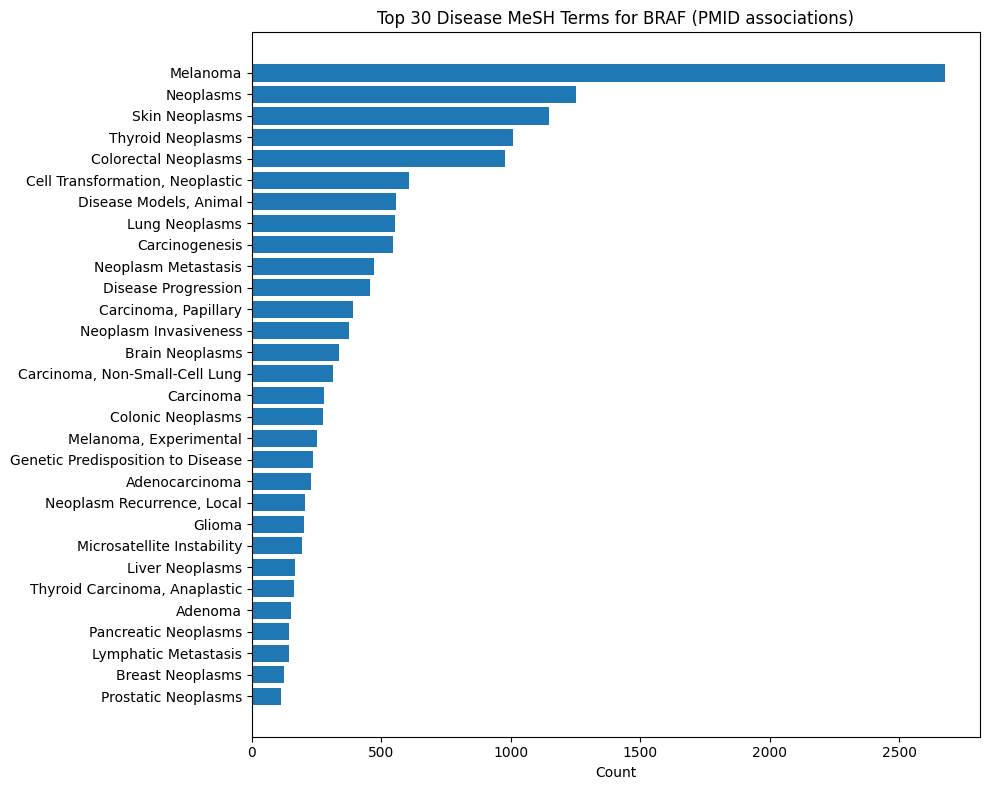
BRAF
INDRA sources:
Colored badges next to statement headings correspond to evidence counts from knowledge sources, as shown below. Badges to the left of the | separator correspond to curated knowledge base sources, and badges to the right of it correspond to machine reading systems.